Research Projects
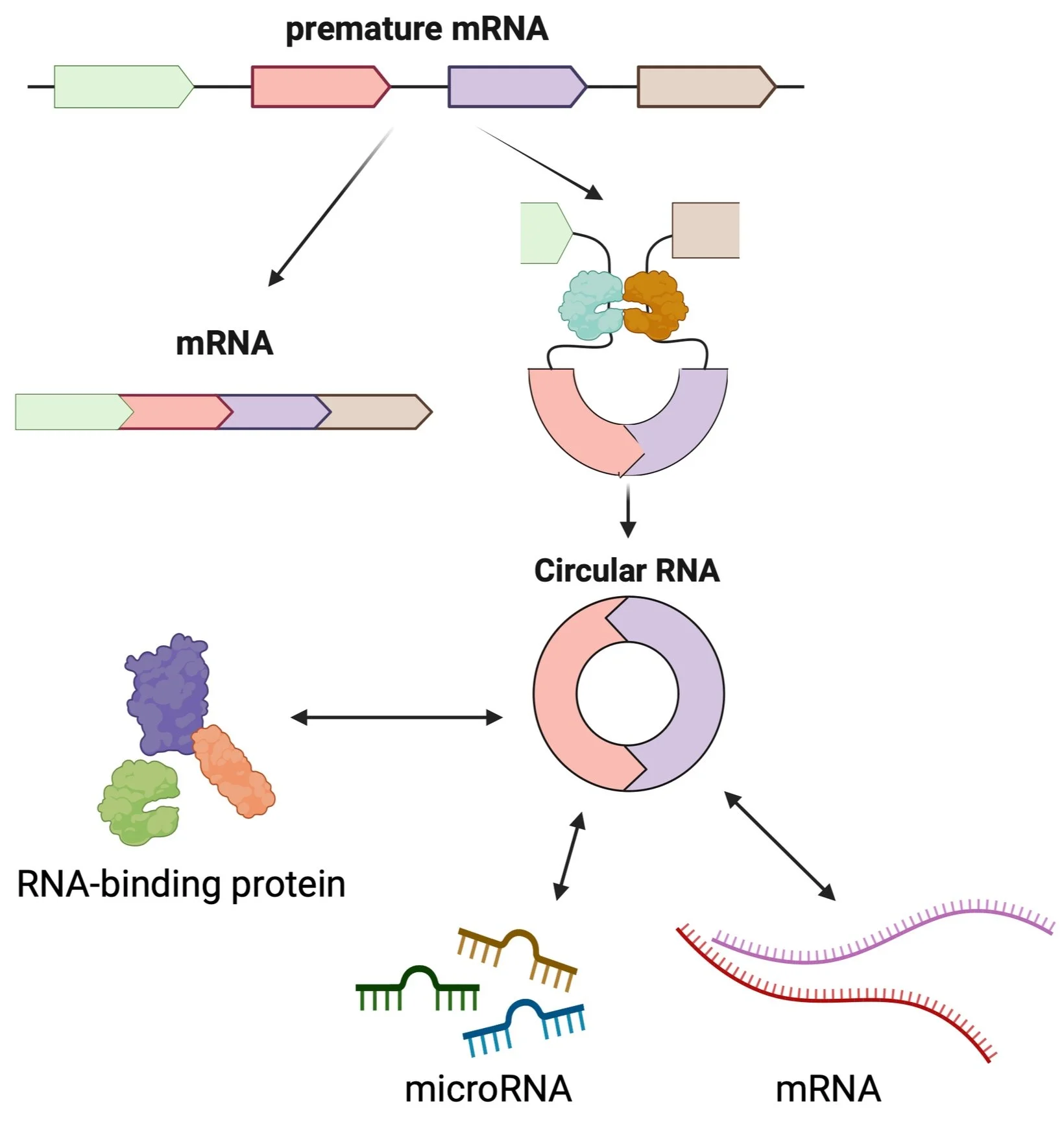
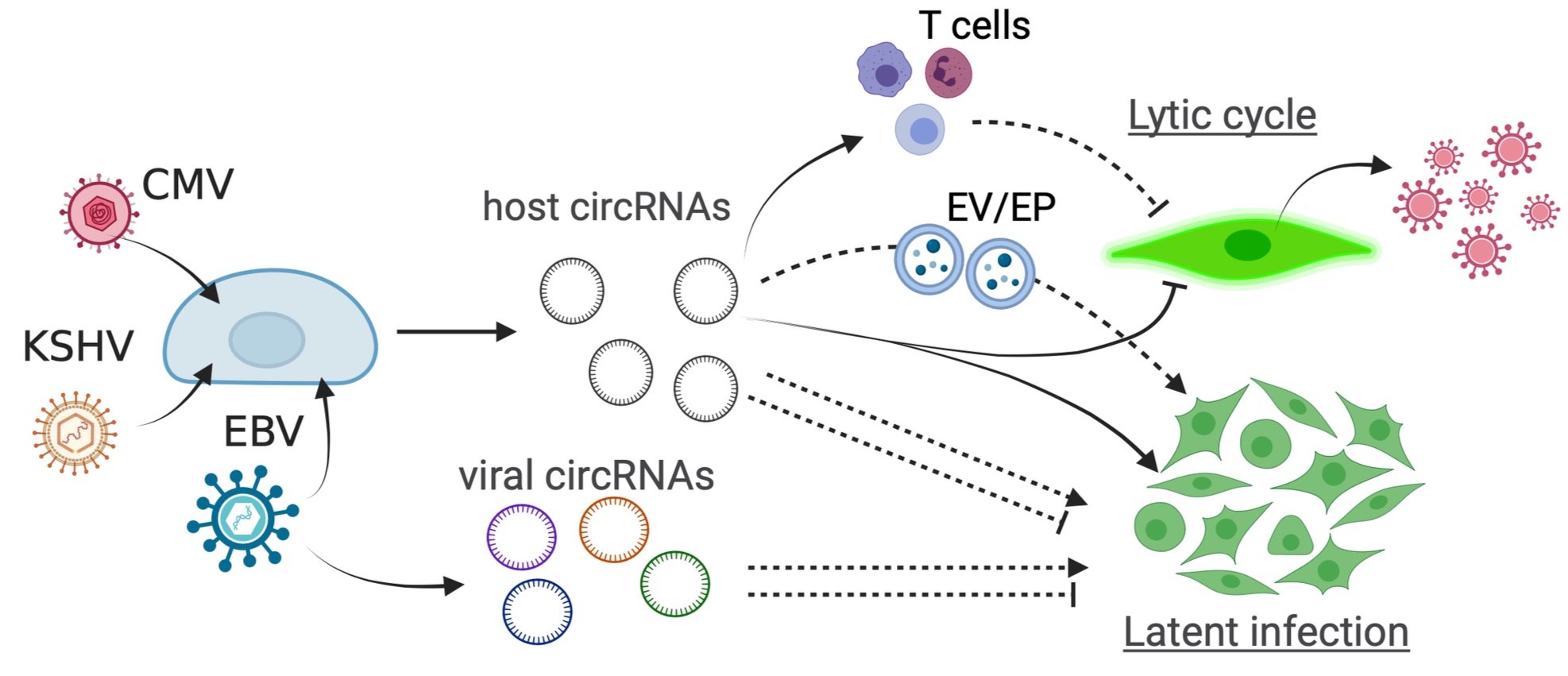
Circular RNA is a novel regulator of virus infection. They are abundant RNA species in human cells and interact with other RNAs and proteins to regulate cellular physiology. The Tagawa lab aims to define functions and mechanisms of non-coding RNAs in virus-host interactions. We are among the first to identify virus infection-induced circular RNAs and to discover herpesvirus-encoded circular RNAs. Using tumour-associated human herpesviruses including Epstein-Barr virus (EBV), Kaposi sarcoma herpesvirus (KSHV), we found a human circular RNA, circRELL1, that can suppress viral lytic cycle, a process that produces new virions and spread infection.
How do human and viral circular RNAs regulate herpesvirus life cycle?
We previously determined host and viral circular RNAs that are induced upon infection with herpesviruses like EBV and KSHV. RNase R exonuclease degrades linear RNA (mRNA or rRNA) but not circular RNAs therefore used to enrich circRNAs. RNA-Seq and microarray were utilized to quantitate circRNAs. We have stringent list of commonly-induced host circRNAs upon EBV/KSHV infection. In this project, we aim to reveal their functions in viral infection, tumorigenesis, and immunity. Combining primary infection models of human herpesviruses (EBV, KSHV, and cytomegalovirus), manipulations of host and viral circular RNAs (RNAi, lentiviral transduction), and high-throughput sequencing techniques, we will interrogate circular RNA’s ability to regulate viral life cycle. Further, we will expand this functional analysis to herpesvirus-encoded circular RNAs. Oncogenic herpesviruses effectively “hide” from host immune surveillance and circular RNAs are ideal candidates to achieve immune evasion thanks to their circular, non-immunogenic nature. This project will thus aim to identify one or more circular RNAs that is crucial for multiple herpesvirus infection.
2. What are impacts of circular RNA on antiviral immunity and tumor microenvironment?
In this project, we aim to reveal how host and viral circular RNAs regulate immune landscapes of (i) infected cells and (ii) neighbouring cells upon infection.
We will first investigate how host and viral circular RNAs effect on abilities of infected cells to evade from antiviral immune cells. RNA-sequencing and immunostaining will be used to identify the targeted immune molecules by circular RNAs such as cell surface molecules and cytokines. Antibody-based neutralization and knock-out studies will determine key immune molecules. Second, circular RNAs are loaded into extracellular vesicles (EVs) and shape the microenvironment. EVs will be purified from infected cells to identify and quantitate of EV-loaded circular RNAs. Further, whether extracellular circular RNAs affect neighbouring cells in innate and adaptive immune gene expression levels will be tested. Manipulation of EV loading and EV secretion will be combined to define contributions of specific human and viral circular RNAs.
3. Which RNAs and proteins are directly bound and regulated by circular RNAs?
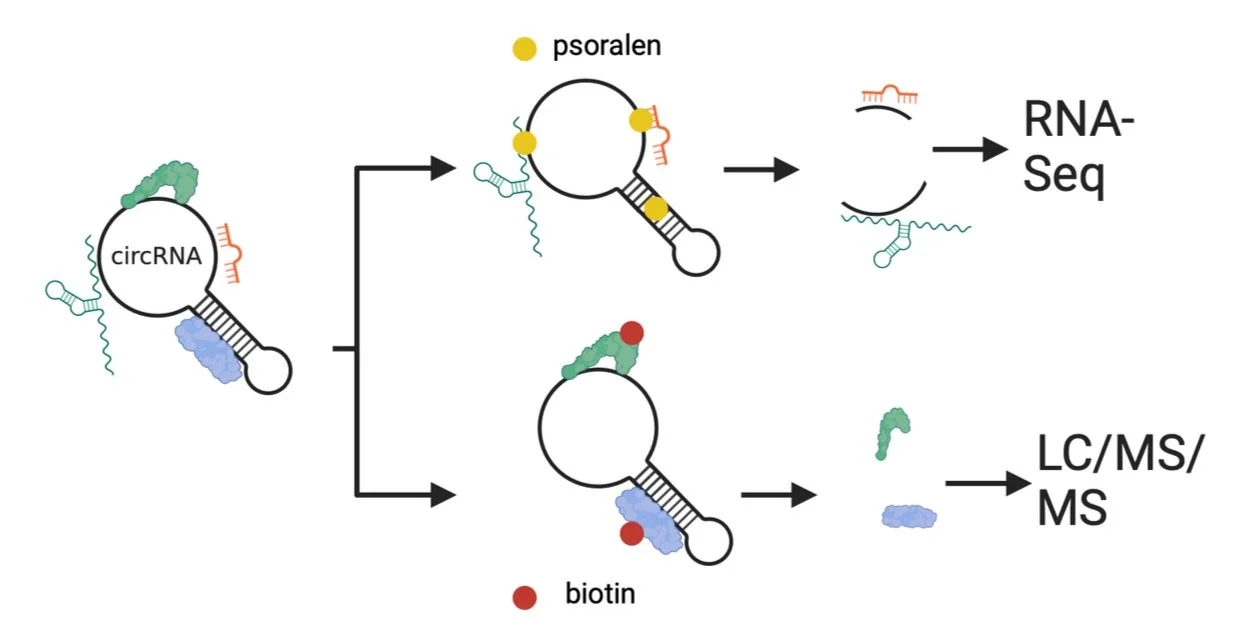
Circular RNAs can interact with other RNAs and proteins to regulate their functions, which may be responsible for the phenotypes caused by circRNAs (aims 1 & 2). Here we will further interrogate mechanisms of host and viral circular RNAs. Previously, we performed circRNA-RNA pulldowns and circRNA-protein pulldowns (below) to reveal the circRELL1 interactome. Psoralen can bridge double-stranded RNA upon UV exposure in reversible manner. circRELL1-RNA interactions were thus covalently bonded, purified, and the bonding was reversed followed by RNA-sequencing. This identified several mRNAs like TTI1 as targets of circRELL1. For proteins, we used CRISPR-Cas13 system to target circRELL1 and purified circRELL1-biding proteins, which were identified with proteome anslysi (LC-MS/MS). Using these techniques, we will investigate targets of commonly-regulated circRNAs by herpesvirus infection and virus-encoded circRNAs. Ecotpic expression and depletion of identified RNAs/proteins will be performed to validate targets and their contributions to circRNA functions.